Research Interests
Organ Size Control and Tumorigenesis
The molecular mechanisms that stop organ growth at the appropriate point during development or regeneration remain unclear. Investigations in the past decades have led to the identification of a number of growth control pathways, including mTOR/Insulin and Hippo signaling pathways that play key roles in regulating multiple events during organism development and homeostasis. Our research is aimed to understand the cellular and molecular mechanisms underlying organ size control and tumorigenesis, with specific emphasis on 1) molecular mechanisms underlying upstream regulation of the Hippo signaling pathway; 2) novel regulation of tissue growth in response to nutrient stress.
1) Molecular mechanisms underlying upstream regulation of the Hippo signaling pathway. The Hippo pathway is an evolutionarily conserved developmental pathway that controls organ size and tissue homeostasis in all metazoan animals. Dysregulated Hippo pathway has been implicated in a wide range of human disorders, including cancer. While the regulation of the core Hippo pathway is well studied, the upstream regulation of this pathway is less defined. A unique feature of these upstream regulators is that their overgrowth phenotypes are relatively mild compared to those of the core components, suggesting the Hippo pathway is regulated by diverse upstream signals. Our previous study identified Kibra-Expanded-Merlin (KEM) protein complex as a key regulatory machinery upstream of the Hippo pathway. Through a large scale genetic screen, our current work aims to identify additional upstream regulators. We anticipate that our work will provide novel mechanistic understanding of how Hippo pathway upstream signals are coordinated.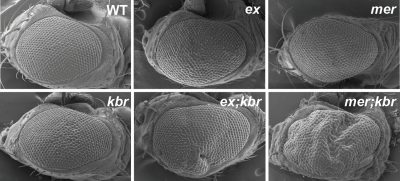
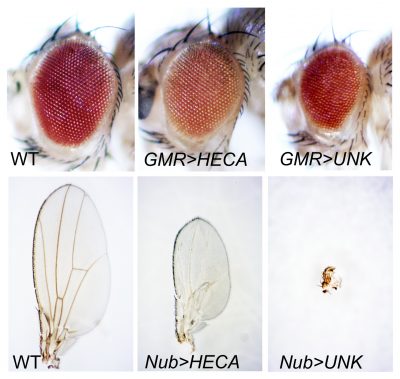
2) Novel regulation of tissue growth in response to nutrient stress. Nutrient restriction (NR) without malnutrition is the most robust environmental intervention that is known to slow a number of diseases (including cancer and aging) in a variety of species. To date, the anti-tumorigenic effects of NR have been well established and its potential implications in both cancer prevention and treatment have been suggested. Despite these advances, our understanding of the molecular mechanisms underlying the anti-tumorigenic effects of NR remain fragmented. We recently identified Hdc and Unk as two NR-specific tumor suppressors. By elucidating how Hdc and Unk activities are regulated by nutrient stress and further, how this regulation determines the sensitive of cells to NR and subsequent NR resistance effects, our research goal is to provide new insights into the molecular mechanisms underlying NR resistance response during animal development, and to have a transformative impact on understanding the development of NR resistance related human diseases.
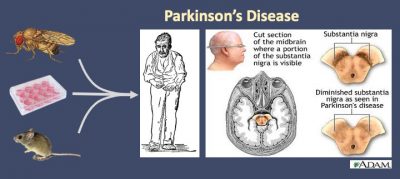
Parkinson’s disease (PD)
Parkinson’s disease (PD) is recognized as the second most common neurodegenerative disorder after Alzheimer’s disease, affecting up to 1% of the population above the age of 60 and 4-5% above the age of 85. The cardinal symptoms are caused by the progressive degeneration of midbrain dopamine (DA) neurons in substantia nigra pars compacta (SNpc) and accompanying striatal pathology and movement deficits. Unfortunately, there is no cure or proven disease modifying therapy for PD. The goal of our research is to understand the molecular mechanisms of PD using Drosophila and mouse systems. As a non-mammalian animal, Drosophila fruit fly has well-defined nerve system, providing a simple yet powerful in vivo system to model neurological diseases. Drosophila adult brain has distinct dopamine (DA) neuronal clusters including about 200 DA neurons. In Drosophila, dysfunction of these DA neurons leads to complicated behaviors mimicking some human PD symposium. As a simple organism, Drosophila also provides great advantages in conducting genome-wide modifier screenings, allowing further identification of components of diverse signaling pathways involved in PD pathogenesis. Taking advantage of the powerful fly genetics, my lab has generated novel transgenic fly models carrying various combinations of human genes with PD-causing mutations (LRRK2, GBA, alpha-Synuclein). By selective expression of these PD genes under dopamine neuron specific driver (TH-Gal4) in combination with environmental stresses, these fly models provide new opportunities to investigate molecular mechanisms underlying PD pathobiology. Particularly, we are trying to understand 1) how do the PD-associated genes cause dopaminergic neurodegeneration, 2) what are the common pathways leading to PD. We expect that the findings from our fly models will be transferred to mammalian systems.